Multimodal Antigenic Escape: GPRC5D T Cell Engagers in Multiple Myeloma
“`html
Differential mutational patterns and convergent evolution of GPRC5D antigen escape clones post anti-GPRC5D TCE
Table of Contents
Patients with relapsed and refractory myeloma progressing on anti-GPRC5D TCE talquetamab (n = 21) were included in this analysis. A summary of the genomic analyses performed on each patient and their clinical characteristics are summarized in Fig. 1a,b and Supplementary Table 1. All patients were treated with talquetamab. All progression CD138+ samples were collected within 1-2 weeks of clinical confirmed disease progression by International Myeloma working Group (IMWG) criteria21. Bulk WGS (100×) was performed on 20 patients (pre-therapy n = 8 and at relapse n = 20). For patients MM-03, MM-10 and MM-20, pre-therapy and relapse scATAC and/or scRNA-seq were performed along with WGS. Samples from patient MM-61 was subjected to targeted sequencing.In terms of clinical response, 14 of 21 cases had at least a very good partial response (VGPR) according to IMWG criteria21. Two patients (MM-65 and MM-67) had primary refractory disease (Fig. 1b). The median progression-free survival (PFS) for the overall group was 12.1 months (95% CI 8-18.1 months) (Fig. 1c).
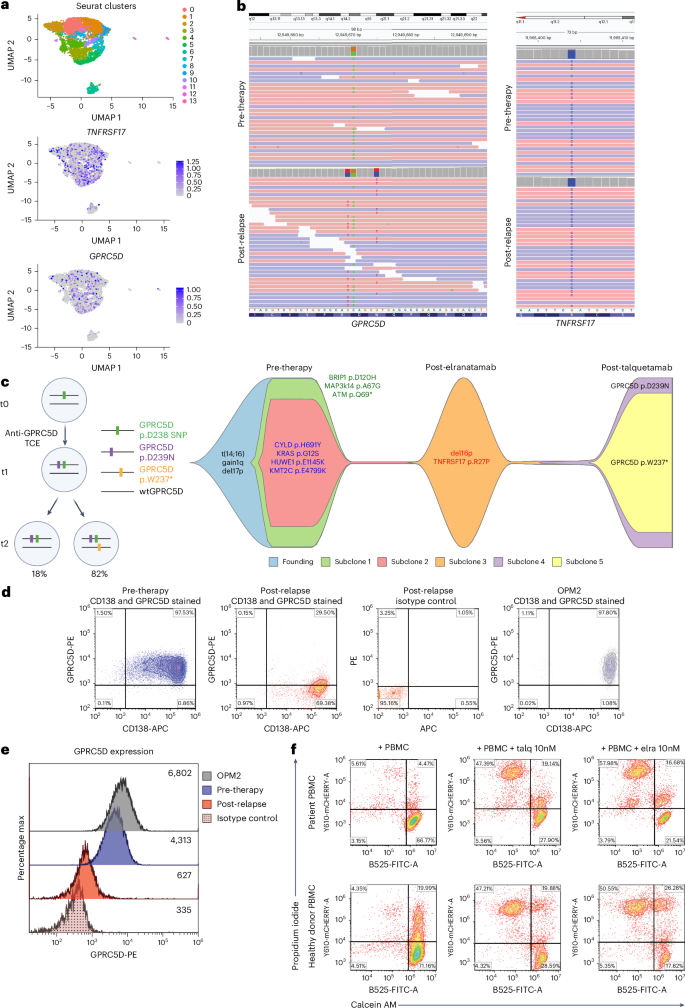
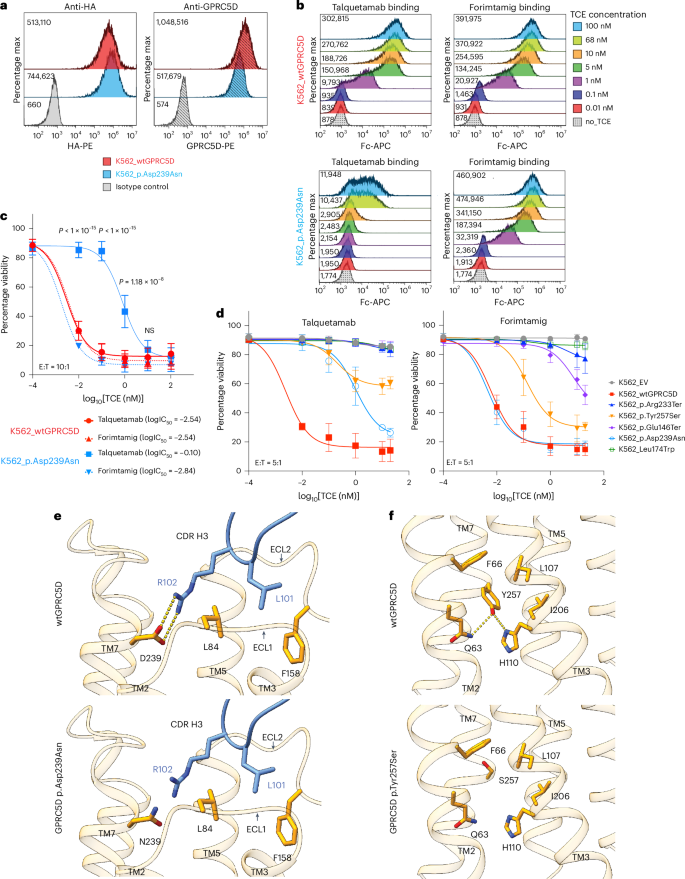
13. CD138+ MM cells at progression harbored monoallelic deletion of TNFRSF17 coupled to a clonal point mutation in the extracellular domain of BCMA (p.Arg27Pro) abrogating anti-BCMA TCE cytotoxicity13. Afterward, the patient was treated with talquetamab, daratumumab and pomalidomide (Talq-DP), and attained stringent complete response (sCR) lasting 13.8 months. scRNA-seq analysis of post-relapse CD138+ MM cells demonstrated retention of GPRC5D transcript (Fig. 2a). Pre-talquetamab WGS revealed a heterozygous germline single-nucleotide polymorphism (SNP) at GPRC5D Asp238 residue (c.714 G > A) (Fig. 1b, top left). At progression, two somatic GPRC5D SNVs were detected by WGS: a clonal missense mutation (p.Asp239Asn, c.715 C > T; cancer cell fractions (CCF) 1) in cis with the Asp238 SNP, and in trans GPRC5D nonsense mutation (p.Trp237Ter, c.710 C > T; CCF 0.82) affecting the second allele (Fig.2b, bottom left). Therefore, as depicted in the phylogenetic tree in Fig. 2b.
a, Uniform manifold approximation and projection (UMAP) of single-cell RNA transcripts (scRNA-seq) data from post-relapse CD138⁺ cells, showing TNFRSF17 (BCMA) and GPRC5D expressions. b, Integrative Genomics Viewer (IGV) snapshots of the GPRC5D and TNFRSF17 loci from pre-treatment and post-relapse CD138⁺ cells (WGS). c, A proposed phylogenetic tree depicting the evolutionary emergence of two GPRC5D mutants identified at progression (t2), with t0 representing pre-therapy and t1 representing an intermediate time point (left). Heterozygous GPRC5D p.D238 SNP is shown in green, in cis GPRC5D p.D239N mutation (purple) and in trans GPRC5D p.W237* mutation (yellow). Created in BioRender. lee, H. (2026) https://BioRender.com/e28w8gr. Fish plot depicting clonal evolution across sequential lines of therapy, with each colored segment representing a genetically distinct subclone (right). therapies administered at each time point are annotated along the timeline. d, Flow cytometry dot plots showing GPRC5D surface expression on CD138⁺ cells pre-treatment and at relapse as well as that of OPM2 MM cells. e, Histogram of flow cytometry results corresponding to d; numbers indicate median fluorescence intensity (MFI). f, Cytotoxicity assay of PBMCs at relapse, co-cultured with OPM2 MM cells at an effector:target (E:T) ratio of 10:1 for 24 h in the presence or absence of 10 nM talquetamab (talq) or elranatamab (elra). Viability of OPM2 MM cells was assessed by calcein AM⁺/propidium iodide⁻ staining. Healthy donor PBMCs were used as a positive control.
Other patients with acquired GPRC5D SNVs included MM-61 with t(11;14) MM who achieved a 12 months complete response on talquetamab. At the time of relapse, targeted amplicon sequencing of the CD138+ tumor cells demonstrated monoallelic deletion
Focal to large biallelic deletion overlapping GPRC5D
A second pattern of GPRC5D antigenic escape resulting from biallelic deletion was observed in five cases. MM-20 received Talq-DP and achieved sCR for 42 months. Bulk WGS of post-relapse tumor demonstrated a chr12p monoallelic deletion with a focal 104-kb biallelic deletion (CCF_CNV 1) encompassing GPRC5D gene locus (chr12: 12,877,450-12,981,981), which were undetectable pre-therapy (Fig. 3a,b). scRNA-seq analysis confirmed loss of GPRC5D transcript in the post-relapse sample and chr12p copy-number loss (inferCNV) (Fig. 3c and Supplementary Fig.2). Flow cytometry was also consistent with nearly absent membrane GPRC5D expression at relapse (Fig. 3d). In vitro cytotoxicity assay using patient PBMCs collected at progression showed efficient killing of OPM2 cells in the presence of 10 nM talquetamab (Fig. 3e). Of note, this patient’s peripheral blood T cell population was predominantly composed of CD8+ T cells (Supplementary Fig.chr12: 12,948,661-12,954,728) (CCF_CNV 0.96) resulting in GPRC5D biallelic deletions, respectively (Extended Data Figs. 5 and 6).
MM-18 demonstrated biallelic GPRC5D deletion (CCF_CNV 0.95) at progression along with a subclonal fraction (CCF 0.04) harboring frameshift deletion GPRC5D p.Leu174TrpfsTer180 (ref. 13). Similarly,MM-19 relapsed with a clonal GPRC5D biallelic deletion (CCF_CNV 1),as we previously reported13.
Epigenetic GPRC5D silencing
The last pattern of acquired resistance involved epigenetic silencing. Patient MM-10 previously received anti-BCMA TCE and progressed after 19 months with a clonal p.Pro34del BCMA mutation13. The patient then received Talq-DP and achieved sCR lasting 19 months. WGS of CD138+ cells at relapse on talquetamab (but not pre-therapy) identified a GPRC5D mutation p.Thr88LeufsTer21 (CCF 0.91) with no other CNVs or structural variants at this gene locus (Fig.
genomic alterations in each tumor sample, including purity, ploidy, variant allelic fractions (VAFs) and purity-corrected cancer cell fractions (ccfs) for SNVs/indels and CNVs, are provided in Supplementary Table 2. Figures summarizing the clonal and subclonal CCF distribution for each patient with antigen escape are included in Supplementary Fig. 7. Two patients, MM-71 and MM-73, achieved sCR with durable remissions lasting 24.8 and 28.1 months,respectively (fig. 1b). At progression, neither patient had evidenAntigen-self-reliant acquired resistance