The working principle of generative artificial intelligence (AI) for new drug development developed by researchers at the Korea Institute of Advanced Science and Technology (KAIST). By learning in advance the interaction patterns between proteins and drug molecules, new drug candidates can be discovered even with a small amount of data./Korea Institute of Advanced Science and Technology
A domestic research team has developed generative artificial intelligence (AI) to help discover and develop new drugs. It is expected that it will be used in the development of new drugs as it can predict the interaction between proteins and new drug candidates even with little knowledge.
Professor Kim Woo-yeon of the Korea Institute of Advanced Science and Technology (KAIST) Department of Chemistry announced on the 18th that they have developed a generative AI that helps develop new drugs with a small amount of data by learning the interaction patterns in advance. between proteins and drug molecules.
Generative artificial intelligence (AI) technology, which creates necessary information based on information input by the user, has been developing rapidly in various fields recently. Like OpenAI’s ChatGPT and Google’s Bard, it helps in everyday life, but is also attracting attention as a next-generation technology in industrial areas such as new drug development.
The development of new drugs begins with finding the protein that causes the disease and the substance that binds to that protein. A new AI-based drug development technology that has recently attracted attention uses generative models to find candidate substances that bind to desired proteins. However, when searching for candidate substances based on the learned data, there is a limitation that results are mostly similar to existing drugs.
This problem has recently been identified as a limitation of new AI-based drug development. In order to treat diseases that cannot be treated with existing drugs, we need to find substances that bind to new proteins, which is difficult due to a lack of experimental data.
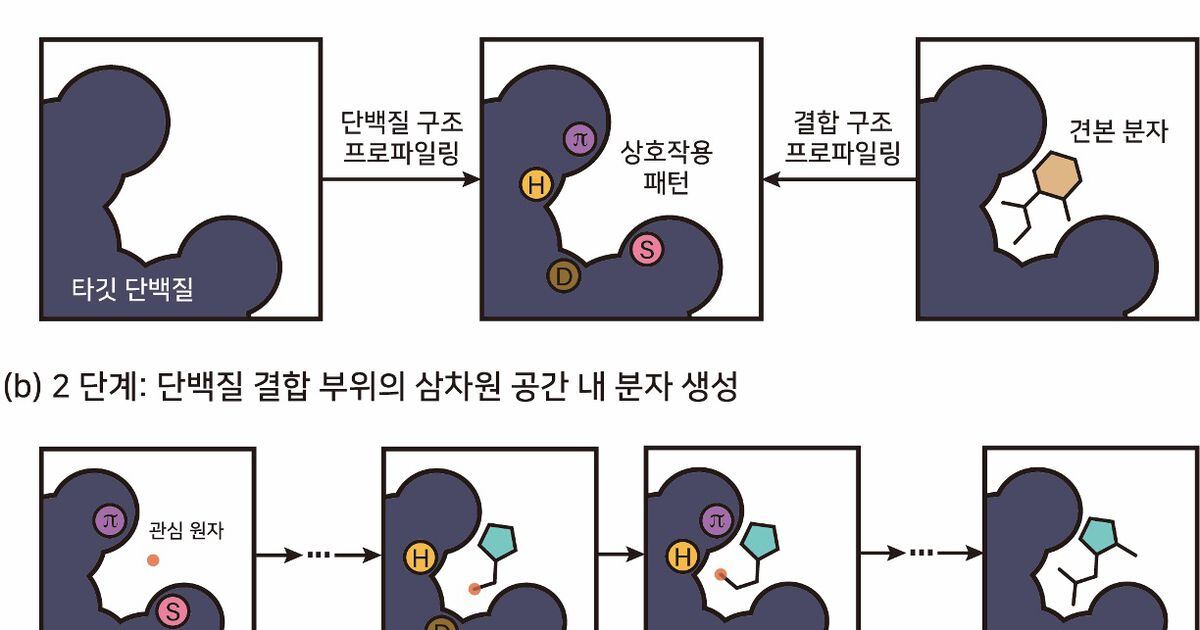
To solve the data dependency problem, KAIST researchers developed a design model to find substances that can only bind with protein structure information. This is a method that uses three-dimensional (3D) information about a protein’s drug-binding site as a template. It is expected that it will be easier to design materials that fit together, just like designing a key that fits into a lock.
The problem of poor stability and cohesion of materials designed with AI models was also solved. To be effective as a new drug, it needs to bind to the desired protein. At the same time, in order to have commercial value as a medicine, it must maintain a stable structure and state of combination within the body.
The researchers noted that the interaction pattern is important when the molecule designed by the AI model binds stably to the protein. We designed a model to learn interaction patterns in generative AI and use them for molecular design.
As a result, it has been shown that it is possible to find new drug candidates by learning thousands of structural data alone. By learning the interaction patterns between proteins and drug molecules using prior knowledge, performance was improved even with small data. Current models require up to 10 million pieces of virtual data to supplement insufficient training data.
Using the newly developed AI, the research team was also able to develop a new drug candidate targeting ‘epithelial growth factor receptor (EGFR)’, which is commonly found in Asian non-small cell lung cancer patients. Mutations in EGFR are known to increase the risk of cancer.
The research team applied the interaction pattern that occurs in the EGFR mutated amino acid molecule to find the binding substance. As a result, it was theoretically shown that 23% of the letters obtained had more than 100 times better binding power than when there was no mutation.
Won-ho Won, a doctoral candidate in the Department of Chemistry at KAIST who participated in this study, said, “Technology that uses prior knowledge is a strategy for active use in fields with relatively little data.” “It will apply to all fields,” he said.
The results of the research were presented on the 15th of last month in the international academic journal ‘Nature Communications’.
Addresses
Nature Communications, DOI:
#finds #drugs.. #Discovery #candidate #substances #lung #cancer #treatment